Research - (2021) Volume 0, Issue 0
Assessment Ukrainian breeds ducks genetic diversity by microsatellite loci
A.M. Chepiha1, S.O. Kostenko2, M.S. Doroshenko2, O.M. Konoval3, L. Lu4, O.V. Sydorenko5*, N.P. Svyrydenko2, P.P. Dzhus5 and M.V. Drahulian6Abstract
Genetic pools of local duck breeds play an essential role as a source of gene polymorphism alleles, which are associated with valuable economic characteristics. Local breeds are represented primarily by Ukrainian White, Ukrainian Clay, Ukrainian Grey, and Ukrainian Black White-Breasted. Two breeds Ukrainian Clay and Ukrainian Black White-Breasted, were studied using a panel of 21 pairs of primers by standard method for DNA isolation, electrophoresis, and statistical analysis. The value of Na in the population of the Ukrainian Clay breed ranged from 2 (CAUD011) to 10 (CAUD050). The highest genetic diversity in the duck population of the Ukrainian Clay breeds was observed by locus CAUD050. According to the results of microsatellite analysis, the number of alleles in the duck population of the Ukrainian Black White-Breasted breed was highest by locus CAUD069. On average, the number of alleles per locus for the Ukrainian Clay breed ducks was 4.429 ± 0.563, while it was slightly higher for the Black White-Breasted breed, i.e., 4.810 ± 0.563. The data obtained can be used to develop biodiversity preservation programs and breeding programs to improve the existing and breed new breeds or crosses of ducks.
Keywords
Allele frequency, locus, population, polymorphic, Ukrainian Clay breed, Ukrainian Black White-Breasted breed.
Introduction
Modern poultry farming includes a wide variety of species, breeds, lines, and crosses of poultry. In 2009, due to the intensification of poultry production, 30% of breeds were threatened with extinction, 9% of breeds were already extinct (Hoffmann,2006), 6% of vanishing breeds are turkeys, 31% are geese, and 25% are ducks. Due to the risk status based on comparable figures from 2006 and 2014, chickens have by far the highest number of breeds at risk among avian species. There are also a few reported cases among ducks, guinea fowl, and turkeys. The regions with the highest proportion of breeds classified as at-risk are Europe and the Caucasus (35 percent of avian breeds). The number of extinct avian breeds by species worldwide: Chicken is 62, Duck 15, Goose 3, Turkey 2 mostly industrial lines that are no longer actively bred. (Hodges, 2006; IUCN Red List, 2006). Preservation of poultry genetic resources is part of institutional activities of many universities in Europe (Özdemir et al., 2016) 26 countries, including eight developing countries, have programs in place to protect the genetic diversity of birds (in situ and ex situ) 24 of those countries have preservation programs for chickens, 7-for ducks, and 2-for geese and turkeys (Sawicka et al., 2011).
In the territory of Ukraine, breeds of waterfowl, particularly ducks, can be found in the form of relatively small local populations. From the subgenus of river ducks, the population of Ukraine is best known for the wild mallard and local domestic species of ducks: Ukrainian White, Ukrainian Clay, Ukrainian Grey, and Ukrainian Black White-Breasted. Therefore, they were used in the 60s of the last century as the initial genetic material when creating domestic national breeds of ducks. Ukrainian Clay breed was developed in the Institute of National Agrarian Academy of Ukraine by pedigree work with local grey ducks. The subsequent selection of birds with clay coats was made. It aimed to maintain typical exterior and bodyweight increasing and, in general, preserve the poultry, fertility, and hatchability.
The compound developed Ukrainian Black White-Breasted breed group reproduced crossbreeding of local black white-breasted ducks with wild Pekin ducks and Khaki Campbell ducks. This breed was predominant among other local ducks with good body weight and typical exterior nature (Ryabokon, 2005; Lyashenko, 2015).
According to the State Breed Registry, as of 01.01.2017, the average adult bird population was 46.6 thousand heads, and as of 2018, there were 38.2 thousand heads. Genetic pools of local duck breeds play an essential role as a source of gene polymorphism alleles associated with valuable economic characteristics (Ostryakova et al., 2010). However, Ukrainian breeds of ducks have not been the subject of systematic genetic research in recent years, so the analysis of their genetic diversity is relevant.
Advances in molecular biology have made it possible to assess the genetic variability of organisms at the DNA level (Weigent et al., 2004; Al-Samarai & Al-Kazaz, 2015). Thus, most researchers use molecular markers to analyze species, populations, or individuals (Yang et al., 2013). In recent years, microsatellites have been actively used to research the genetic diversity of different breeds and populations of ducks in the world (Yinhua et al., 2005; Wu et al., 2009; Agatep et al., 2016; Seo et al., 2016; Carcò et al., 2018; Hariyono et al., 2019). The priority of use of microsatellite loci in researching the genetic variability of populations is because they allow obtaining molecular genetic information for a detailed characterization of polymorphism of species and breeds (Marzanov et al., 2011; Zinovyeva and Gladyr, 2011; Fisinin et al., 2011; Novgorodova et al., 2012; Tao et al., 2016; Fisinin et al., 2017). In recent years, an increasing number of microsatellites have been developed for waterfowl (Maak et al., 2000; Genet et al., 2003; Huang et al., 2005; Seo et al., 2016). A genetic map of duck, along with the use of microsatellite loci, will allow to the research of biological diversity and genetic relations between different breeds of this species (Maak et al., 2003; Liu et al., 2006; Wang et al., 2006; Huang et al., 2009).
Thus, due to the lack of genetic information about Ukrainian breeds of ducks, it was decided to analyze the genetic diversity of the Ukrainian Clay and Ukrainian Black White-Breasted breeds by microsatellite DNA loci.
Materials and Methods
The subject of the research was ducks of two breeds: Ukrainian Clay (n=9) of Povit-Agro Farming Enterprise, Kyiv region, and Ukrainian black white-breasted (n=11) of Zdolbunivske Private Agricultural Enterprise of Breeding Poultry Farming, Rivne region.
Duck venous blood samples were collected in tubes containing 3 ml of EDTA. According to the manufacturer's instructions, DNA was extracted using the Sorb-B commercial kit (AmpliSense, Russia).
Amplification of microsatellite loci was performed on a 2720 Cycler Gene Amp PCR thermocycler (Applied Biosystems Inc., USA) using the following program: denaturation at 94°C for 3 minutes, 35 cycles of denaturation at 94°C for 15 seconds, annealing at 50–68°C (depending on the primer, Table 1) for 15 seconds, elongation at 72°C for 30 seconds, final elongation at 72°C for 3 minutes (Tao et al., 2016).
| Locus | The sequence of primers (5’–3') | Repeated sequence | Annealing temperature, °C |
|---|---|---|---|
| APL11 | F: AACTACAGGGCACCTTATTTCC R: TTGCATCAGGGTCTGTATTTTC |
(GA)25 | 60 |
| APL12 | F: AGTTGACCCTAATGTCAGCATC R: AAGAGACACTGAGAAGTGCTATTG |
(GA)27 | 60 |
| APL26 | F: AACAGGGATAACATGAGAAGTGG R: TGAGCAGCTGTCTGGTATCTATTC |
(CA)11(GA)9 | 55 |
| APL80 | F: ggatgttgcc ccacatattt R: ttgccttgtt tatgagccat TA |
(AT)4(GT)11 | 58 |
| APL79 | F: acatctttgg cattttgaa R: catccactag aacacagaca TT |
(TTCC)18 | 55 |
| APL78 | F: aaccaagaca gaataatcct ta R: gaacacaact gctttgcta |
(GT)9(AT)5 | 55 |
| CMO12 | F: ggatgttgcc ccacatattt R: ttgccttgtt tatgagccat t |
(AT)15 | 58 |
| CMO11 | F: ctccactaga acacagacat t R: catctttggc attttgaag |
(GGAA)13(GGGA)15 | 45 |
| APH01 | F: TACCTTGCTCTTCACTTTCTTT R: GTATGACAGCAGACACGGTAA |
(CA)10 | 47 |
| APH09 | F: GGATGTTGCCCCACATATTT R: TTGCCTTGTTTATGAGCCATTA |
(CA)11 | 55 |
| APH10 | F: ATTAGAGCAGGAGTTAGGAGAC R: GCAAGAAGTGGCTTTTTTC |
(CA)12 | 55 |
| SMO7 | F:TTTTCACCCAGTTCACTTCAGCC R:GATTCAAATTTGCCGCAGGATTA |
(GT)12 | 55 |
| SMO11 | F:AAATCAACCAAAGAGGCATAGCC R:GCAGTTGTTTTGGAGGACAGACA |
(TG)12GA(G)13(AG)5 | 68 |
| SMO12 | F:CCTGGTGGGATAGGTTTAAAATG R:TGTTCATCAAAAGCAGAGAGGGG |
(TG)9T11 | 47 |
| SMO13 | F:ACCATCTTCCTTTCCTCCCAACC R:GGGCTTGAGGCATACACTCCCTA |
(TG)13(AC)2(TG)2 | 58 |
| APH04 | F-CCTGCTGCCTTCCACAACACT R- GTGCTGACCGTCATGGTGCAG |
(AC)15 | 61 |
| CAUD069 | F-CTCATTCCAATTCCTCTGTA R-CAGCATTATTATTTCAGAAGG |
(TTCC)3TTTC(TTCC)5 (TTTC)9CTTC(TTTC)18 |
52 |
| CAUD011 | F-CAAAGTTAGCTGGTATCTGC R-TGCTATCCACCCAATAAGTG |
(CA)13 | 53 |
| CAUD049 | F-TGTAGTTTAGTTGCTGGATA R-TTAGTAAACTCTTGCCATCT |
(TTTC)8TTCC(TTTC)17 | 50 |
| CAUD050 | F-GGACAAGTGGCATGTGTCAT R-GGCTTCTGTGCTCCTCAGAT |
(TCTCTTTC)9…(TTTC) | 59 |
| CAUD019 | F-CTTAGCCCAGTGAAGCATG R-GCAGACTTTTACTTATGACTC |
(TTTC)23 | 55 |
Table 1. Description of primers of microsatellite loci.
The total volume of the reaction mixture was 15 µL:1.5 µL 10 × Buffer, 1 µL DNA, 1.5 µL Mg2+(25 mmol/L), 0.3 µL (10 mmol/L) dNTP, 0.3 µL (5 U/µL) Taq enzyme, locus–specific primers (upstream and downstream) of 0.15 µL each (10 nmol), deionized H2O–11.6 µL (GenetBio, Korea). The genotyping mixture contained 1 µL PCR product, 10 µL Hi-Di formamide and 0.1 µL GeneScan-500LIZ size standard (Applied Biosystems, USA) (Tao et al., 2016).
Electrophoretic analysis of the amplification products was conducted on an automatic four-capillary genetic analyzer 3130 DNA Analyzer (Applied Biosystems Inc., USA). The graphic data obtained was decoded and documented on a computer to automatically decode DNA fragmentation analysis results of Genotyper® and Gene Mapper ™.
Statistical analysis was performed using conventional methods (Zhivotovskiy, 1991). The number of alleles (Na) was calculated using the GenAlEx 6.5 software (Peakall & Smouse, 2006; Peakall & Smouse, 2012).
Results and Discussion
Microsatellite analysis demonstrated that the population's four loci of the Ukrainian Clay breed were monomorphic: APH10, SMO7, SMO12, SMO13. The total number of identified alleles of Ukrainian Clay ducks by 17 polymorphic loci was 89 (Fig. 1). APL11, CAUD019, CAUD050 loci were identified in 8 individuals of Ukrainian Clay breed from 9 ones researched. According to the research results, the value of Na in the population of the Ukrainian Clay breed ranged from 2 (CAUD011) to 10 (CAUD050). The following loci were most polymorphic: APH09, APL11, APL12, APL26, APL80, CAUD019, CAUD049, CAUD050, CAUD069, CMO11, and CMO12. The number of alleles at those loci ranged from 5 (APL12, APL26, APL80, CAUD019) to 10 (CAUD050) alleles per locus.
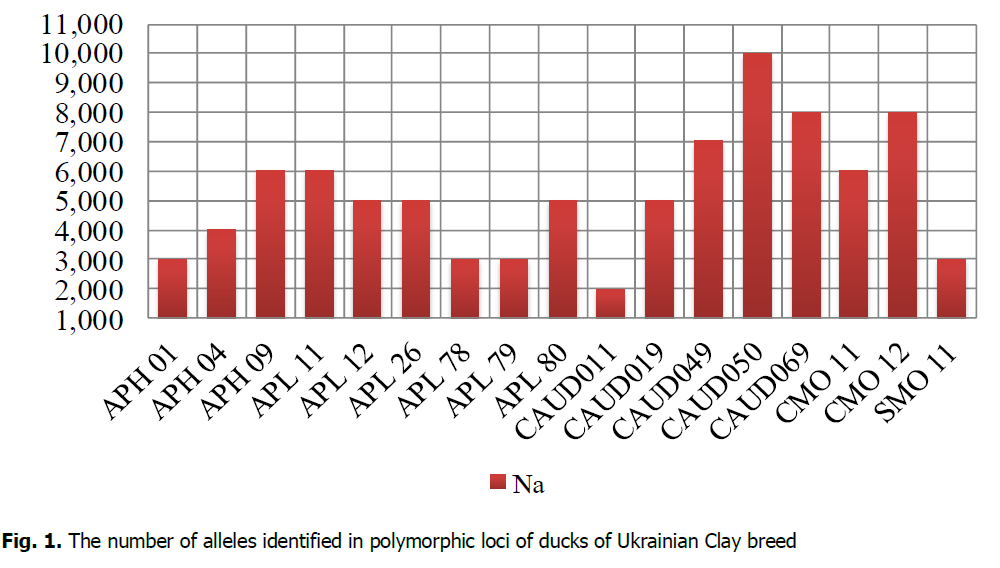
Figure 1: The number of alleles identified in polymorphic loci of ducks of Ukrainian Clay breed.
The analysis of the genetic diversity of ducks in the population of the Ukrainian Black White-Breasted breed was performed by 21 microsatellite DNA loci. The research established that the APH10 locus was monomorphic. The total number of alleles identified in the population of Ukrainian Black White-Breasted was 100 (Fig. 2) by 20 polymorphic loci. APL26, APL78, APL80, CAUD049, CAUD069, SMO11 loci were identified in 10 individuals of Ukrainian Black White-Breasted breed from 11 ones researched.
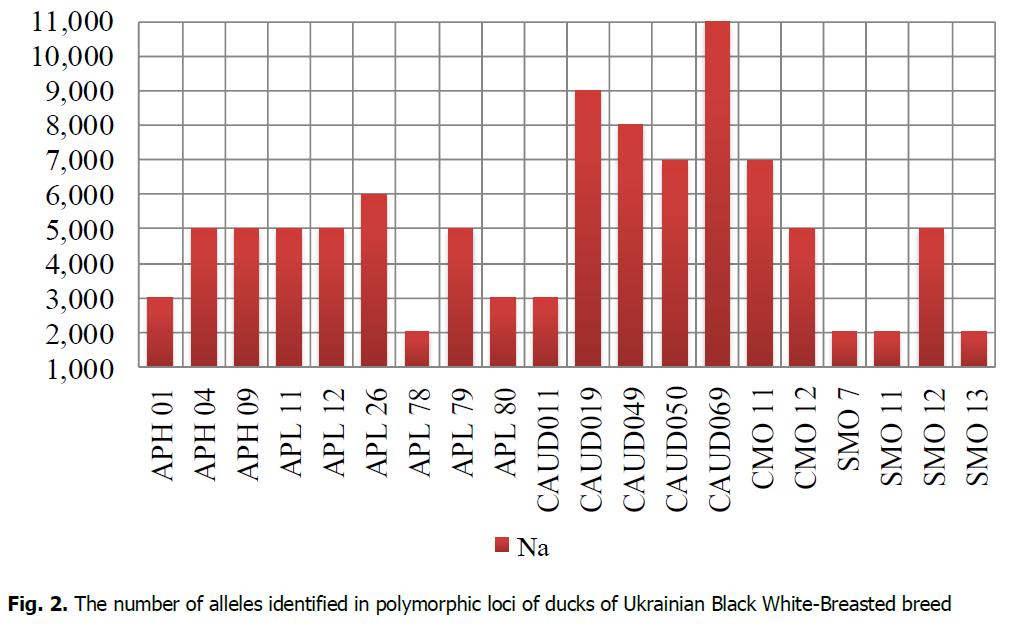
Figure 2: The number of alleles identified in polymorphic loci of ducks of Ukrainian Black White-Breasted breed.
Following microsatellite analysis, the highest number of ducks alleles in the Ukrainian Black White-Breasted breed population was identified for CAUD069 locus (11 alleles), and the smallest–for APL78 SMO7, SMO11, and SMO13 (2 alleles per locus). The following loci were most polymorphic: APH04, APH09, APL11, APL12, APL26, APL79, CAUD019, CAUD049, CAUD050, CAUD069, CMO11, CMO12, and SMO12. The number of alleles at those loci ranged from 5 (APH04, APH09, APL11, APL12, APL79, CMO12, SMO12) to 11 (CAUD069). According to Ahmadi et al. (2007), SMO7 and SMO13 loci of Peking and Muscovy ducks were also monomorphic. According to Agatep et al. (2016), different duck breeds were characterized because seven microsatellite loci of 28 researched could be both mono- and polymorphic. The difference in the number of alleles (Na) and their frequencies in polymorphic loci make it possible to establish genetic diversity and evolutionary processes within and between populations (Hillel et al., 2007).
According to the results of our research, ducks of the Ukrainian Clay breed demonstrated the highest number of alleles by locus CAUD050, while ducks of the Ukrainian Black White-Breasted breed demonstrated the highest number of alleles by locus CAUD069. Our data confirm Carco et al. (2018) results, which testify to polymorphism and a significant number of alleles in loci CAUD050 and CAUD069 if ducks of Italian and Polish breeds (Carco et al., 2018). Seo et al. (2016) also established a high degree of polymorphism by the number of alleles of locus CAUD069 of duck populations in South and East Asia (Seo et al. 2016). Thus, given the polymorphism in populations of different origins, loci CAUD050 and CAUD069 can be recommended for use in the certification of individual duck lines and families as universal.
The highest genetic diversity in the Ukrainian Clay breeds duck population was observed by locus CAUD050 (Fig. 3). The frequencies of alleles in this locus in the duck populations researched varied from 0.063 (CAUD 050246, CAUD 050308, CAUD 050372, CAUD 050380, CAUD 050424) to 0.250 (CAUD 050300) in the population Ukrainian Clay breed, while the minimum frequency rate in the population of Ukrainian Black White-Breasted breed was 0.045 (CAUD 050302, CAUD 050380) and the maximum frequency rate was 0.318 (CAUD 050290). Allelic variants of CAUD050300, CAUD050302, CAUD050304, CAUD050312 and CAUD050380 were common for ducks of the Ukrainian Clay and Ukrainian Black White-Breasted breeds. However, ducks in the populations of the Ukrainian Clay breed had alleles in locus CAUD050 that were characteristic only of this breed and were not found at ducks of the Ukrainian Black White-Breasted breed: CAUD050246, CAUD050308, CAUD050324, CAUD050372, CAUD050424. In particular, ducks of the population of Ukrainian Black White-Breasted had alleles CAUD050290, CAUD050306, which were not found in the population of the Ukrainian Clay breed.
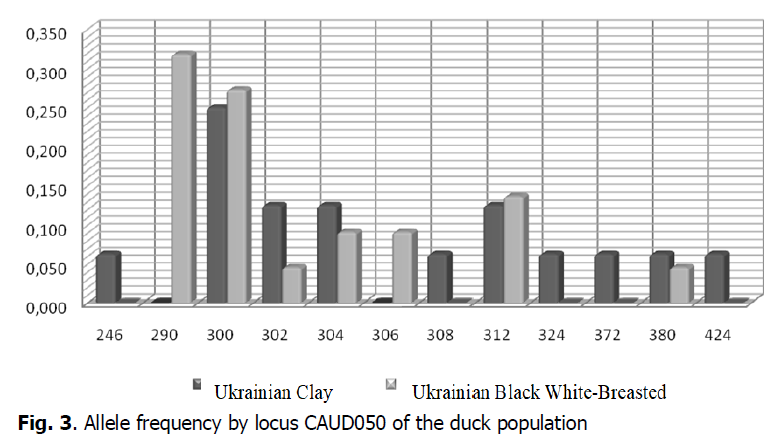
Figure 3: Allele frequency by locus CAUD050 of the duck population
According to the results of microsatellite analysis, the number of alleles in the duck population of the Ukrainian Black White-Breasted breed was highest by locus CAUD069 (Fig. 4). The allele frequency rate per locus was maximum by CAUD069196 (0.200), and the minimum allele frequency rate was 0.050 (CAUD069200, CAUD069206, CAUD069216, CAUD069250, CAUD069254). The allele frequency rate per locus of duck of the population of the Ukrainian Clay breed ranged from 0.056 (CAUD069192, CAUD069210, CAUD069258, CAUD069262) to 0.278 (CAUD069184, CAUD069200). Alleles CAUD069184, CAUD069188, CAUD069192, CAUD069200, and CAUD069258 were common by locus CAUD069 for ducks of the populations researched. The highest number of alleles by locus CAUD069 was identified for ducks of the Ukrainian Black White-Breasted breed (11 alleles); they included those that were not found in this locus in the Ukrainian Clay breed population: CAUD069180, CAUD069196, CAUD069206, CAUD069216, CAUD069250, CAUD069254. According to the results of microsatellite analysis, ducks in the Ukrainian Clay breed population had alleles that were characteristic of this breed and were not found in populations of the Ukrainian Black White-Breast bread: CAUD069201, CAUD069210, CAUD069262.
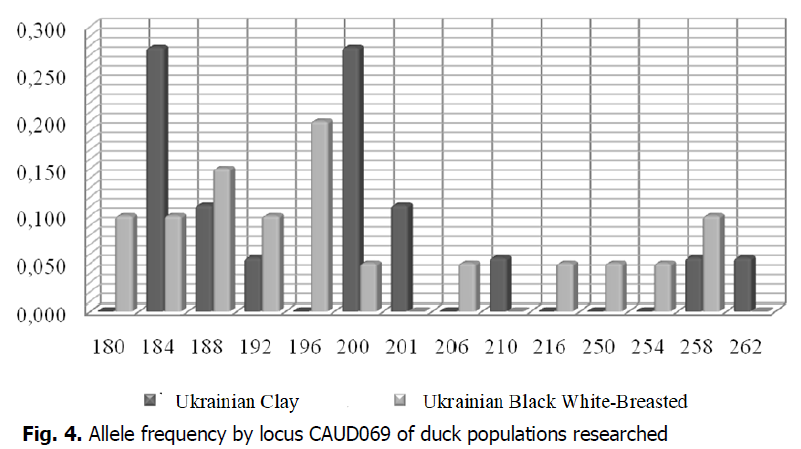
Figure 4: Allele frequency by locus CAUD069 of duck populations researched.
The data obtained indicate a genetic similarity of the analysed local breeds of ducks of Ukrainian breeding by those loci, but the presence of alleles found only in a separate population indicates the differences in the genetic profile of ducks.
On average, the number of alleles per locus for the Ukrainian Clay breed ducks was 4.429 ± 0.563, while it was slightly higher for the Black White-Breasted breed, i.e., 4.810 ± 0.563. This indicator indicates a relatively high level of genetic diversity of the duck breeds researched. Thus, according to Ahmadi et al. (2007), it was found that the total number of alleles per locus was 2.2 for Peking ducks and 2.44 Muscovy ducks on average. In the research of Agatep et al. (2016), this indicator did not exceed 2.714 for ducks of three populations (Peking, Khaki Campbell, and Philippine duck).
According to the results of our research, ducks of Ukrainian breeds have a higher number of alleles on average per locus as compared to modern populations of source rocks (Peking and Khaki Campbell). This indicates a higher genetic diversity of modern populations of domestic breeds than modern breeds whose ancestors were representatives of the parental breed. The phenomenon of high polymorphism of specific loci of the breeds researched can be due to several reasons: polymorphism of the source material (parental breed could be more polymorphic than their current populations); crossing systems (inbreeding, outbreeding, interlinear crossing) and selection; representativeness of the samples analyzed; polymorphism of loci used for analysis.
Effective identification of breeds and species and the genetic uniqueness of breeds depends on the presence of microsatellite loci characterized by a large number of rare alleles. Unfortunately, the majority of results of duck research obtained in different laboratories are presented arbitrarily without identifying alleles, which makes it impossible to compare genotypes of different populations and certify individual lines. Thus, there is a need to unify approaches to the analysis of the genetic diversity of ducks with due regard to the results of previous research of peculiarities of polymorphism of highly polymorphic loci.
Following our research, we identified alleles for ducks of Ukrainian breeds, which were specific to a particular population only. Thus, ducks in the population of the Ukrainian Clay breed (Table 2) had the highest number of them by locus (CAUD050246, CAUD050308, CAUD050324, CAUD050372, CAUD050424) with a frequency of 0.063 alleles per locus. The maximum allele frequency rate was identified by locus APH04 locus (APH04142)–0.611. The average frequency of alleles characteristic of ducks in the Ukrainian Clay breed population, which were not identified in the Ukrainian Black White-Breasted breed, was 0.107.
| Locus | Alleles | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|
| CAUD050 | 5 | 246 | 308 | 324 | 372 | 424 |
| APH09 | 3 | 108 | 117 | 194 | ||
| CAUD069 | 3 | 201 | 210 | 262 | ||
| CMO12 | 3 | 108 | 110 | 118 | ||
| APL80 | 2 | 108 | 118 | |||
| CAUD049 | 2 | 284 | 288 | 246 | ||
| CMO11 | 2 | 109 | 239 | |||
| APH 01 | 1 | 203 | ||||
| APH04 | 1 | 142 | ||||
| APL11 | 1 | 120 | ||||
| APL12 | 1 | 156 | ||||
| APL26 | 1 | 143 | ||||
| APL76 | 1 | 215 | ||||
| CAUD019 | 1 | 130 | ||||
| SMO11 | 1 | 206 |
Table 2. Specific alleles of ducks in the Ukrainian Clay population
According to the research results, the population of ducks of the Ukrainian Black White-Breasted breed had alleles that were characteristic of this breed and were not found at the polymorphic loci of the ducks of the Ukrainian Clay breed (Table 3). The highest number of alleles was identified by locus CAUD069 (CAUD069180, CAUD069196, CAUD069206, CAUD069216, CAUD069250, CAUD069254). The allele frequency rate ranged from 0.050 (CAUD069206, CAUD069216, CAUD069250, CAUD069254) to 0.200 (CAUD069196). The highest frequency rate was identified by locus CAUD050 (CAUD050290–0.318). The average frequency of specific alleles for the duck population of the Ukrainian Black White-Breasted breed was 0.077.
| Locus | Alleles | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|
| CAUD069 | 6 | 180 | 196 | 206 | 216 | 250 | 254 |
| CAUD019 | 5 | 134 | 160 | 190 | 198 | 214 | |
| SMO12 | 4 | 64 | 88 | 130 | 164 | ||
| CAUD049 | 3 | 244 | 268 | 280 | |||
| CMO11 | 3 | 219 | 235 | 255 | |||
| APH04 | 2 | 140 | 144 | ||||
| APH09 | 2 | 124 | 196 | ||||
| APL26 | 2 | 148 | 152 | ||||
| APL79 | 2 | 224 | 240 | ||||
| CAUD050 | 2 | 290 | 306 | ||||
| APH01 | 1 | 197 | |||||
| APL12 | 1 | 146 | |||||
| CAUD011 | 1 | 130 | |||||
| SMO7 | 1 | 181 | |||||
| SMO13 | 1 | 190 |
Table 3. Specific alleles of ducks in the Ukrainian Black White-Breasted population
The research results concerning identifying specific alleles in the duck breeds analyzed should be considered from a small sample of individuals. With increasing the number of poultry researched, the number of breed-specific alleles will be smaller. However, because of the decreasing number of populations of local duck breeds in Ukraine (Ryabokon et al., 2005), future research should consider the data obtained to preserve the genetic diversity of Ukrainian duck breeds, their certification, and identification.
Conclusion
Following analysis of the data on the genetic diversity of ducks of the Ukrainian breeds obtained, the most polymorphic loci were identified for the Ukrainian Clay breed (APH09, APL11, APL12, APL26, APL80, CAUD019, CAUD049, CAUD050, CAUD069, CMO11, CMO12) and the Ukrainian Black White-Breasted breed (APH04, APH09, APL11, APL12, APL26, APL79, CAUD019, CAUD049, CAUD050, CAUD069, CMO11, CMO12, SMO12) based on microsatellite DNA analysis. We found the population of the Ukrainian Black White-Breasted breed was more polymorphic by the number of alleles (4.810 ± 0.563 alleles per locus) compared to the population of the Ukrainian Clay breed (4.429 ± 0.563 alleles). The highest number of alleles was identified by locus CAUD050 (10) in the Ukrainian Clay breed ducks. In the Ukrainian Black White-Breasted ducks population, locus CAUD069 was the most polymorphic-11 allelic variants. The data obtained can be used to develop biodiversity preservation programs and breeding programs to improve the existing and breed new breeds or crosses of ducks.
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding
This study was supported by the Earmarked Fund for National Waterfowl-industry Technology Research System (CARS-42-06) and the Zhejiang Major Scientific and Technological Project of Agricultural (livestock's) Breeding (grant number 2016C02054-12).
References
Agatep, R.C., Lambio, A.L., Vega R.S.A., Capitan, S.S., Mendioro, M.S., Yebron, M.G.N. (2016). Microsatellite-based genetic diversity and relationship analyses of three genetic groups of domesticated mallard ducks (Anas platyrhynchos domesticus L.). Philippine Journal of Veterinary and Animal Sciences, 42:102–111.
Ahmadi, A.K., Rahimi, G., Vafaei, A., Sayyazadeh, H. (2007). Microsatellite analysis of genetic diversity in Pekin (Anas platyrhynchos) and Muscovy (Cairina moschata) duck populations. International Journal of Poultry Science, 6:378-382.
Alloui, N., Tlidjene, M., Alloui-Lombarkia, O., Zeghina, D. (2003). 2003 Spring meeting of the WPSA French Branch. British Poultry Science, 44:771-772.
Al-Samarai, F.R., Al-Kazaz, A.A. (2015). Molecular markers: An introduction and applications. European Journal of Molecular Biotechnology, 9:118-130.
Carcò, G., Grajewski, B., Cassandro, M., Lisowski, M., Szwaczkowski, T. (2018). Genetic variability of some Italian and Polish duck breeds. Italian Journal of Animal Science, 17:899-906.
Chang-Wen, H., Yu-Shin, C., Rouvier, R., Kuo-Tai, Y., Wu, C.P., Huang, H.L., Mu-Chiou, H. (2009). Duck (Anas platyrhynchos) linkage mapping by AFLP fingerprinting. Genetics, Selection, Evolution, 41:1.
Fisinin, V.I., Gladyr, E.A., Volkova, V.V., Sevast'yanova, A.A., Zinovyeva, N.A. (2011). Assessment of genetic structure of chicken breeds using microsatellite markers. Problemyi biologii produktivnyih zhivotnyih, 1:68–72.
Fisinin, V.I., Selionova, M.I., Shinkarenko, L.A., Shcherbakova, N.G., Kononova, L.V. (2017). Study of microsatellites in the Russian breeds of turkey. Biology Agricultural, p:739.
Hariyono, D.N.H., Maharani, D., Cho, S., Manjula, P., Seo, D., Choi, N., Lee, J.H. (2019). Genetic diversity and phylogenetic relationship analyzed by microsatellite markers in eight Indonesian local duck populations. Asian-Australasian Journal of Animal Sciences, 32:31-37.
Hillel, J., Granevitze, Z., Twito, T., Ben-Avraham, D., Blum, S., Lavi, U., Weigend, S. (2007). Molecular markers for the assessment of chicken biodiversity. World's Poultry Science Journal, 63:33-45.
Hodges, J. (2006). Conservation of genes and culture: historical and contemporary issues. Poultry science, 85:200-209.
Hoffmann, I. (2009). Open questions on poultry genetic diversity. In World Poultry Science Association (WPSA), 6th European Poultry Genetics Symposium, Bedlewo, Poland, pp:61-73.
http://www.redlist.org/.
Huang, Y., Tu, J., Cheng, X., Tang, B., Hu, X., Liu, Z., Li, N. (2005). Characterization of 35 novel microsatellite DNA markers from the duck (Anas platyrhynchos) genome and cross-amplification in other birds. Genetics Selection Evolution, 37:1-18.
Huang, Y., Tu, J., Cheng, X., Tang, B., Hu, X., Liu, Z., Li, N. (2005). Characterization of 35 novel microsatellite DNA markers from the duck (Anas platyrhynchos) genome and cross-amplification in other birds. Genetics Selection Evolution, 37:1-18.
Liu, W., Hou, Z.C., Huang, Y.H., XU, G., Qu, L., Yao, J., Yang, N. (2006). Genetic relationship among Chinese domestic ducks as revealed by microsatellite analysis. In Proceeding of the 30th International Conference on Animal Genetics.
Lyashenko. Yu.V. (2015). Ocinka rivnya genetychnoyi minlyvosti u vitchyznyanyx porodnyx grupax siryx ta glynyastyx kachok z vykorystannyam RAPD-markeriv. Suchasne Ptaxivnycztvo, 10:16–18.
Maak, S., Neumann, K., Lengerken, G.V., Gattermann, R. (2000). First seven microsatellites developed for the Peking Duck (Anas platyrhynchos). Animal Genetics.
Maak, S., Wimmers, K., Weigend, S., Neumann, K. (2003). Isolation and characterization of 18 microsatellites in the Peking duck (Anas platyrhynchos) and their application in other waterfowl species. Molecular Ecology Notes, 3:224-227.
Marzanov, N.S., Devrishov, D.A., Marzanova, S.N., Komkova, E.A., Ozerov, M.Yu., Kantanen, Yu. (2011). Genetic labeling, conservation of biodiversity and the problems in animals breeding. Agricultural Biology, 2:3–14.
Novgorodova, I.P., Fisinin, V.I., Mikhajlov, M.V., Rojter, Y.S., Brem, G. (2012). Investigation of information value of the multiplex test systems for analysis of chicken with the different number of loci. Research and Technical Advances of Agribusiness Sector.
Ostryakova, O.E., Gadyuchko, O.T., Katerynych, O.O. (2010). Identification of the gene pool porids for the main loci of poultry feathering. Mizhvidomchiy tematichniy zbirnik, Ptahivnitstvo, 65:57–68.
Özdemir, D., Maretto, F., Cassandro, M. (2016). Comparison of genetic diversity of Turkish and Italian local chicken breeds for further conservation strategies. European Poultry Science, 80:1-14.
Peakall, R., Smouse, P.E. (2012). GenAlEx tutorials-part 2: genetic distance and analysis of molecular variance (AMOVA). Bioinformatics, 28:2537-2539.
Peakall, R.O.D., Smouse, P.E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6:288-295.
Riabokon, Yu.O., Pabat, V.O., Mykytiuk, D.M., Frolov, V.V., Katerynych, O.O., Bondarenko, Yu.V., Mosiakina, T.V., Hadiuchko, O.T., Kovalenko, H.T., Bohatyr, V.P., Liutyi, Yu.S. (2005). Catalon breeding resources in the sil'kohospodar bird of Ukraine/pid. Kharkiv, p:78.
Sawicka, D., Brzezinska, J., Bednarczyk, M. (2011). Cryoconservation of embryonic cells and gametes as a poultry biodiversity preservation method. Folia Biologica (Kraków), 59:1-5.
Seo, D., Bhuiyan, M.S.A., Sultana, H., Heo, J.M., Lee, J.H. (2016). Genetic diversity analysis of South and East Asian duck populations using highly polymorphic microsatellite markers. Asian-Australasian Journal of Animal Sciences, 29:471.
Tao, Z., Xu, X., Shen, J., Li, L., Zeng, T., Dong, S., Lu, L. (2016). Analysis of genetic diversity and relationship among 6 wild duck breeds and Shaoxing partridge duck (Anas platyrhynchos domestic). Journal of Agricultural Biotechnology, 24:1173-1180.
Wang, L.G., Yu, D.B., Du, W.X. (2006). Microsatellite analysis of genetic diversity between Beijing duck and Cherry Valley duck. Jiangsu Agricultural Science, 6:299–301.
Weigend, S., Romanov, M. N., & Rath, D. (2004). Methodologies to identify, evaluate and conserve poultry genetic resources. World's Poultry Congress, Istanbul, Turkey.
Wu, F., Huang, Y., Ma, Y., Hu, S., Hao, J., Li, N. (2009). Evaluation of genetic diversity and relationships within and between two breeds of duck based on microsatellite markers. Progress in Natural Science, 19:1581-1586.
Yang, W., Kang, X., Yang, Q., Lin, Y., Fang, M. (2013). Review on the development of genotyping methods for assessing farm animal diversity. Journal of Animal Science and Biotechnology, 4:1-6.
Zhivotovskiy, L.A. (1991). Populjacionnaja biometrija. M.:Nauka, p:270.
Zinovieva, N.A., Gladyr, E.A. (2011). Geneticheskaya ekspertiza sel'skokhozyaystvennykh zhivotnykh: Primenenie test-sistem na osnove mikrosatellitov [Genetic evaluation of agricultural animals: The use of microsatellite test system]. Dostizheniya Nauki i Tekhniki APK, 9:19-20.
Author Info
A.M. Chepiha1, S.O. Kostenko2, M.S. Doroshenko2, O.M. Konoval3, L. Lu4, O.V. Sydorenko5*, N.P. Svyrydenko2, P.P. Dzhus5 and M.V. Drahulian62National University of Life and Environmental Sciences of Ukraine, 12 Heroiv Oborony Str., Kyiv, 03041, Ukraine
3Laboratory of Quality and Safety of Product of Agro-Industrial Complex of Ukraine of the National Un, Kyievo-Sviatoshynskyi District, Kyivska Region, 08162, Ukraine
4Institute of Animal Husbandry and Veterinary Science of Zhejiang Academy of Animal Sciences, 198 Shiqiao Road, Hangzhou, 310021, China
5Institute of Animal Breeding and Genetics named after M.V.Zubets of National Academy of Agrarian Sci, Chubynske, Boryspil District, Kyivska Region, 08321, Ukraine
6RBW Genetik GmbH, 88339, Bad Waldsee, Germany
Citation: Chepiha, A.M., Kostenko, S.O., Doroshenko, M.S., Konoval, O.M., Lu, L., Sydorenko, O.V., Svyrydenko, N.P., Dzhus, P.P., Drahulian, M.V. (2021). Assessment Ukrainian breeds ducks genetic diversity by microsatellite loci. Ukrainian Journal of Ecology, 11 (6), 105-112.
Received: 26-Jul-2021 Accepted: 09-Aug-2021 Published: 23-Aug-2021
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
