Research - (2021) Volume 0, Issue 0
Analysis of the potential associated with the siderophores synthesis in the Bacillus subtilis strain using whole genome sequencing
T.P. Dunyashev1*, G.Yu. Laptev1, E.A. Yildirim2, L.A. Ilina2, V.A. Filippova2, D.G. Tiurina2, A.V. Dubrovin2, N.V. Tarlavin2, S.N. Bikonya2, E.A. Brazhnik2, V.H. Melikidy2 and A.V. Platonov3Abstract
The use of biological products based on beneficial microorganisms that produce siderophores, which reduce the concentration of available to pathogensiron ions, can be successfully used in agriculture. The aim of our study was the sequencingand functional annotation of the genome of Bacillus subtilis strain from the collection of BIOTROF Ltd to identify genes associated with the synthesis of siderophores. Nucleotide sequences were analyzed using a MiSeq instrument (Illumina, Inc., USA) together with a MiSeq Reagent Kit v3 (300-cycle) (Illumina, Inc., USA). Functional genome annotation was carried out using the PROKKA 1.12 (https://github.com/kbaseapps/ProkkaAnnotation http://vicbioinformatics.com/) and RAST 2.0 (https://rast.nmpdr.org) programs. The KEGG Pathway database was used to assess the pool of genes associated with antimicrobial activity and construct a metabolic map (http://www.genome.jp/kegg/). When carrying out bioinformatic processing of whole genome sequencing data of the B. subtilis strain, several gene clusters associated with the synthesis of siderophores were identified. Аlmost all genes necessary for the implementation of the process of iron binding with the participation of bacillibactin were found. A cluster of DhbA, DhbB, DhbC genes associated with the production of enzymes 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28), isochorismatase (EC 3.3.2.1) and isochorismates synthase (EC 5.4.4.2), which are responsible for synthesis of bacillibactin precursor was found. The cluster of genes FeuA, FeuB, FeuC, and FeuD is associated with the synthesis of substrate-binding proteins of the iron transport system. In the genome of B. subtilis, genes associated with the synthesis of other siderophores were also found-enterochelin, described for E. coli and Campylobacter coli, alsomyxochelin A, first found in Angiococcus disciformis. It is likely that B. subtilis bacteria can synthesize several synergistic siderophores to increase their competitiveness. In our opinion, there is a potential for using the B. subtilis strain in animal husbandry and plant growing as a basis for bacterial biopreparations-producers of siderophores, in order to reduce the pathogenic load.
Keywords
Bacillus subtilis, siderophores, whole genome sequencing, probiotics, PGPR.
Introduction
Many bacteria require metal ions, in particular, ironions, which carries out the functions of electron transfer and be a cofactor in the synthesis of DNA and RNA, to ensure their vital activity (Capdevila D.A., et al., 2017). Soluble Fe3+ ions are usually present in the environment (water, soil, etc.) in very low concentrations (Harris W.R., 2002). Iron homeostasis in the mammalian body is also tightly regulated; almost all iron ions in blood serum are associated with heme and proteins (ferritin, transferrin, etc.) (Andrews S.C., et al., 2003). Therefore, during the evolution microorganisms have formed specific molecular structures-siderophores-low molecular weight chelating agents that allow bacteria to assimilate iron ions in a bound state (Johnstone T.C., et al., 2015). Siderophores differ in structure and chemical properties; according to the functional groups of metal chelation, they can be classified as hydroxamates, catecholates, carboxylates, and mixed types (Khan А., et al., 2018).
The highest efficiency of siderophore synthesis is characteristic of pathogenic microorganisms and usually correlates with their virulence level (Abaturov A.E., et al., 2018; Koehler T.M., 2000). Controlling the presence of iron in mammals serves as a barrier to infection (Andrews S.C., et al., 2003). It has been established that an excess of iron in microorganisms is associated with the development of a chronic course of the infectious process, since high levels of iron ions contribute to the formation of biofilms of pathogenic bacteria (Banin E., et al., 2005). There is evidence that the use of probiotic preparations based on beneficial microorganisms that produce siderophores, which reduce the concentration of iron ions available to pathogens, can have a positive effect on the health of animals and birds (Balakrishna A., et al., 2012).
Siderophore producers are also used in plant growing (De Serrano Luis O., 2017). Due to the production of siderophores and successful competition for iron ions present in the soil, PGPR -rhizobacteria can inhibit phytopathogenic microorganisms (Rout M.E., et al., 2013). On the other hand, plant endo- and epiphytes synthesizing siderophores can contribute to improving the absorption of iron from the soil by plants, and also activate the production of the growth hormone indole-3-acetic acid (Rout M.E., et al., 2013).
Nevertheless, the pathways for biosynthesis of drugs based on siderophores are laborious, and the yield of the aimed product is low. Therefore, the use of bacterial biological products based on producers of siderophores, as natural biocontrol agents in animal husbandry and crop production, can be a very promising direction (Havenaar R., et al., 1992; Gatesoupe F.J., 1999).
The aim of the research was the sequencing and functional annotation of the Bacillus subtilis strain genome in order to identify genes associated with the synthesis of siderophores that are important for use in agriculture.
Materials and Methods
The material for the study was the B. subtilis strain from the collection of BIOTROF Ltd, which has a significant biocontrol activity. DNA isolation was carried out by standard methods using the Genomic DNA Purification Kit (Fermentas, Inc., Lithuania) according to the attached instructions. A DNA library for whole genome sequencing was prepared using the Nextera XT kit (Illumina, Inc., USA). Nucleotide sequences were determined using a MiSeq instrument (Illumina, Inc., USA) together with a MiSeq Reagent Kit v3 (300-cycle) (Illumina, Inc., USA). Invalid sequences and adapters were removed using the Trimmomatic-0.38 program (https://www.osc.edu/book/ex-port/html/4385) (Bolger A.M., et al., 2014). Filtered in length from 50 to 150 bp pair-terminal sequences were assembled de novo using the SPAdes-3.11.1 genomic assembler (http://cab.spbu.ru/software/spades/). Functional genome annotation was carried out using the PROKKA 1.12 (https://github.com/kbaseapps/ProkkaAnnotation http://vicbioinformatics.com/) and RAST 2.0 (https://rast.nmpdr.org) programs (Aziz R.K., 2008). The KEGG Pathway database (http://www.genome.jp/kegg/) was used to assess the pool of genes associated with antimicrobial activity and construct a metabolic map (Kanehisa M., et al., 2012).
Results
During bioinformatics processing of whole genome sequencing data of B. subtilis strain, several gene clusters associated with the synthesis of siderophores, such as Bacillibactin, Mixochelin and Enterochelin (Enterobactin, Enterobacterin), were identified.
Table 1 shows the genes found in the genome of the studied B. subtilis strain, necessary for the transport of iron, as well as the biosynthesis of Bacillibactin. For example, a cluster of DhbA, DhbB, DhbC genes associated with the production of enzymes 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28), isochorismatase (EC 3.3.2.1) and isochorismates synthase (EC 5.4.4.2). The genes for the assembly of bacillibactin DhbE, DhbF and a cluster of genes of the iron transport system: FeuA, FeuB, FeuC, and FeuD were also found.
| S.No. | Gene Name | Enzymes Associated With Genes |
|---|---|---|
| 1 | DhbA | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28) |
| 2 | DhbB | Isochorismatase (EC 3.3.2.1) |
| 3 | DhbC | Isochorismatessynthase (EC 5.4.4.2) |
| 4 | DhbE | 2,3-dihydroxybenzoate-AMP ligase (EC 2.7.7.58) |
| 5 | DhbF | Component F of bacillibactin synthetase (EC 2.7.7.) |
| 6 | YuiI | Trilactonehydrolase |
| 7 | Feu | Regulatory component: Aa iron transport system(substrate-binding protein) |
| 8 | FeuA | Iron transport system A (substrate-binding protein) |
| 9 | FeuB | Iron transport system B (substrate-binding protein) |
| 10 | FeuC | Iron transport system C (substrate-binding protein) |
| 11 | FeuD | Iron transport system D (substrate-binding protein) |
Table 1. Genes and enzymes of the B. subtilis bacterial strain associated with the synthesis of Bacillibactin (using the RAST database (https://rast.nmpdr.org)).
Figure 1 shows metabolic synthesis maps (according to the KEGG Pathway database (http://www.genome.jp/kegg/)) of some siderophores, in particular, bacillibactin, in the studied B. subtilis strain. According to the obtained results (Fig. 1), the precursors for the biosynthesis of siderophores in B. subtilis are formed from the pool of amino acids in a simple way. In the first step, the resulting chorismate is isomerized to isochorismate by the enzyme chorismate-hydroxymutase (EC 5.4.4.2). Further, isochorismatase (EC 3.3.2.1) catalyzes the chemical reaction of the conversion of isochorismate to pyruvate and 2,3-dihydroxy-2,3-dihydroxybenzoate, which is a precursor in the biosynthesis of these siderophores. Then the enzyme complex enterobacterin synthase (N-(2,3-dihydroxybenzoyl) -serine synthetase) (EC 6.3.2.14) catalyzes the conversion of three molecules of 2,3-dihydroxybenzoate and L-serine into siderophore enterochelin. This complex is formed with the participation of the EntB and EntE genes. The EntE protein catalyzes the ATP-dependent condensation of 2,3-dihydroxybenzoate and the EntB protein to form the covalently arylated form of EntB. The MxcE and MxcF proteins, in turn, participate in the assembly of the myxochelin siderophore from lysine and 2,3-dihydroxybenzoic acid with the participation of ATP and NAD(P)H.
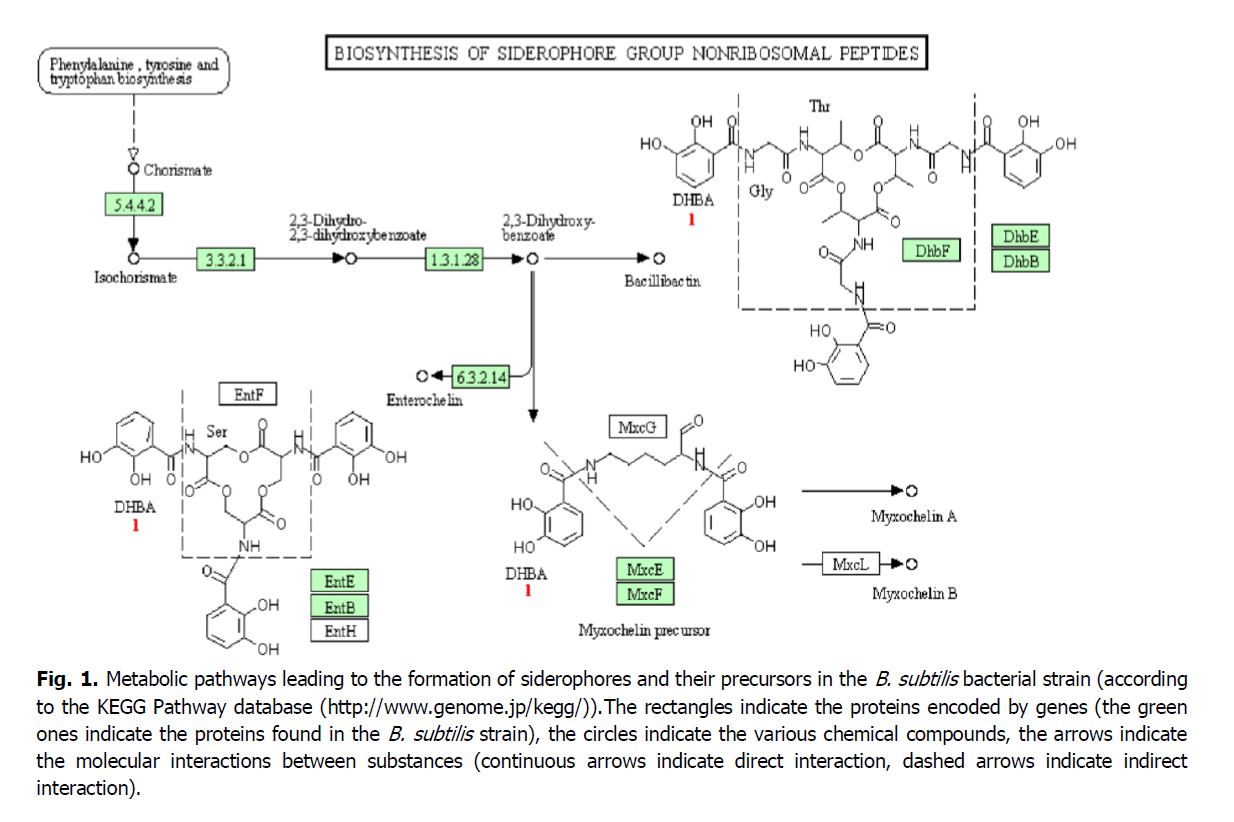
Fig. 1. Metabolic pathways leading to the formation of siderophores and their precursors in the B. subtilis bacterial strain (according to the KEGG Pathway database (http://www.genome.jp/kegg/)).The rectangles indicate the proteins encoded by genes (the green ones indicate the proteins found in the B. subtilis strain), the circles indicate the various chemical compounds, the arrows indicate the molecular interactions between substances (continuous arrows indicate direct interaction, dashed arrows indicate indirect interaction).
Discussion
The results of our study showed that in the genome of the B. subtilis strain, almost all genes necessary for the process of iron binding with the participation a bacillibactin siderophore were found. Siderophore bacillibactin is a catechol trilactone secreted by various types of bacilli during periods of reduced concentration of iron ions in the medium (Hotta K., et al., 2010). A quantitative assessment of intracellular and extracellular bacillibactin in the Bacillus subtilis strain revealed a fourfold increase in the concentration of this extracellular siderophore in an environment with a low content of iron ions compared to its increased content (Miethke M., et al., 2006). The identified cluster of genes DhbA, DhbB, DhbC is associated with the synthesis of enzymes that are responsible for the synthesis of the bacillibactin precursor. Other discovered genes-DhbE, DhbF-are involved in the assembly of bacillibactin. The cluster of genes FeuA, FeuB, FeuC, and FeuD is associated with the synthesis of substrate-binding proteins of the iron transport system (Heinrichs D.E., et al., 2004). Miethke et al. showed that the feuABC gene locus, encoding ABC transporters, including the periplasmic binding protein FeuA and membrane permeases FeuB and FeuC, was of decisive importance for maintaining cell growth of B. subtilis at a low concentration of iron ions in the nutrient medium. The YuiI gene is associated with the process of iron release from bacillibacterial chelate in the cytosol (Miethke M., et al., 2008).
Interestingly, genes associated with the synthesis of other siderophores-enterochelin (enterobactin), described for E. coli (Langman L., et al., 1972) and Campylobacter coli (Richardson Peter T., et al., 1995), and alsomyxochelin A, first described for Angiococcusdisciformis (Myxobacterales) (Kunze B., et al., 1989) were found (Fig. 1). Genes associated with the synthesis of secondary metabolites, including siderophores, are most prone to horizontal transfer (Ziemert N., et al., 2014). The identification in the genome of B. subtilis of genes associated with the synthesis of enterochelin and myxochelin A, previously found in unrelated bacteria, is a clear demonstration of evolutionary mechanisms action in relation to secondary metabolites. The persistence of these genes and even entire clusters in the genome indicates a high degree of their functionality, since their preservation is associated with an increase in genome size and replication costs. In the absence of their functionality, selection pressure would lead to the deletion of "excess genes". Probably, B. subtilis bacteria can synthesize several siderophores, acting in synergism, to increase their competitiveness, which was previously shown for other microorganisms as well (Barona-Gómez F., et al., 2006). In B. cereus, transporter proteins specific for bacillibactin and enterobactin (FeuA, FatB, FpuA, YfiY, etc.) were identified (Zawadzka A.M., et al., 2009).
This is a very important scientific and practical result, since bacterial siderophores are of great interest for their use in plant growing and animal husbandry. Previously, it was shown that treatment of wheat plants with some species of the genus Bacillus producing siderophores had a positive effect on an increase in the mass of roots and aerial parts (Ravari S.B., et al., 2014). Research by Yu, X.M., et al., demonstrated that treatment of pepper plants with a siderophore-producing bacterial strain Bacillus subtilis suppressed fusarium wilt and stimulated plant growth. Bacillibactin produced by Bacillus subtilis AH18 was considered as a substance with the potential to be used in agriculture as a biocontrol agent (Woo S.M., et al., 2008).
Conclusion
Thus, we carried out bioinformatic processing of the data of whole genome sequencing of the B. subtilis strain and identified several clusters of genes associated with the synthesis of siderophores. Thus, almost all genes necessary for the implementation of the process of iron binding with the participation of bacillibactin were found. A cluster of genes DhbA, DhbB, DhbC associated with the production of enzymes 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase, isochorismate and isochorismates synthase, which are responsible for the synthesis of the bacillibactin precursor, was revealed. The cluster of genes FeuA, FeuB, FeuC, and FeuD is associated with the synthesis of substrate-binding proteins of the iron transport system. In the genome of B. subtilis, genes associated with the synthesis of other siderophores were also found-enterochelin, described for E. coli and Campylobacter coli, as well as myxochelin A, first found in Angiococcus disciformis. It is likely that B. subtilis bacteria can synthesize several siderophores, acting in synergism, to increase their competitiveness, which was previously shown for other microorganisms as well.
In the future, there is a need for additional studies that can compare the predicted molecular characteristics with the empirically observed biochemical and physiological patterns of siderophore synthesis in microorganisms. To formulate fundamental conclusions and practical application of siderophore producers, various methods of detecting and determining the characteristics of these substances (liquid chromatography, mass spectrometry, nuclear magnetic resonance spectroscopy) should be used. Further efforts to investigate the synthesis of siderophores by microorganisms will help uncover the potential of their use in agriculture.
Funding
The study was supported by the RFBR grant No. 19-316-90041 "Whole-genome sequencing of bacilli strains isolated from the ruminal contents of various ruminants".
References
Capdevila, D.A., Edmonds, K.A., Giedroc, D.P. (2017). Metallochaperones and metalloregulation in bacteria. Essays in biochemistry, 61:177-200.
Harris, W.R. (2002). Iron chemistry. Molecular and cellular iron transport. Marcel Dekker, New York (NY), p:1-35.
Andrews, S.C., Robinson, A.K., Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiology Reviews, 27:215-237.
Johnstone, T.C., Nolan, E.M. (2015). Beyond iron: Non-classical biological functions of bacterial siderophores. Dalton Transactions, 44:6320-6339.
Khan, A., Singh, P., Srivastava, A. (2018). Synthesis, nature and utility of universal iron chelator–Siderophore: A Review. Microbiological Research, 212:103-111.
Abaturov, A.E., Kryuchko, T.A. (2018). Drug restriction of the availability of iron ions for pathogenic bacteria (part 1). Child Health-Child Health, 13:416-424.
Koehler, T.M. (2000). Gram-positive pathogens. Washington, DC: American Society for Microbiology. Banin, E., Vasil, M.L., Greenberg, E.P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proceedings of the National Academy of Sciences, 102:11076-11081.
Balakrishna, A., Kumar, N.A. (2012). Preliminary studies on siderophore production and probiotic effect of bacteria associated with the Guppy, Poecilia reticulata Peters, 1859. Asian Fish Science, 25:193-205.
De Serrano, L.O. (2017). Biotechnology of siderophores in high-impact scientific fields. Biomolecular Concepts, 8:169-178.
Rout, M.E., Chrzanowski, T.H., Westlie, T.K., DeLuca, T.H., Callaway, R.M., Holben, W.E. (2013). Bacterial endophytes enhance competition by invasive plants. American Journal of Botany, 100:1726-1737.
Havenaar, R., Ten Brink, B., Huis, J.H. (1992). Selection of strains for probiotic use. In Probiotics, p:209-224.
Gatesoupe, F.J. (1999). The use of probiotics in aquaculture. Aquaculture, 180:147-165. Bolger, A.M., Lohse, M., Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30:2114-2120.
Aziz, R.K., Bartels, D., Best, A.A., DeJongh, M., Disz, T., Edwards, R.A., Zagnitko, O. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics, 9:1-15.
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research, 40:109-114.
Hotta, K., Kim, C.Y., Fox, D.T., Koppisch, A.T. (2010). Siderophore-mediated iron acquisition in Bacillus anthracis and related strains. Microbiology, 156:1918-1925.
Miethke, M., Klotz, O., Linne, U., May, J.J., Beckering, C.L., Marahiel, M.A. (2006). Ferri‐bacillibactin uptake and hydrolysis in Bacillus subtilis. Molecular Microbiology, 61:1413-1427.
Crosa, J.H., Mey, A.R., Payne, S.M. (2004). Iron transport in bacteria. Washington, DC: ASM press.
Miethke, M., Schmidt, S., Marahiel, M.A. (2008). The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. Journal of Bacteriology, 190:5143-5152.
Langman, L., Young, I.G., Frost, G.E., Rosenberg, H., Gibson, F. (1972). Enterochelin system of iron transport in Escherichia coli: mutations affecting ferric-enterochelin esterase. Journal of Bacteriology, 112:1142-1149.
Richardson, P.T., Park, S.F. (1995). Enterochelin acquisition in Campylobacter coli: Characterization of components of a binding-protein-dependent transport system. Microbiology, 141:3181-3191.
Kunze, B., Kohl, W., Hofle, G., Reichenbach, H. (1985). Production, isolation, physico-chemical and biological properties of angiolam A, a new antibiotic from Angiococcus disciformis (Myxobacterales). The Journal of Antibiotics, 38:1649-1654.
Ziemert, N., Lechner, A., Wietz, M., Millán-Aguiñaga, N., Chavarria, K.L., Jensen, P.R. (2014). Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proceedings of the National Academy of Sciences, 111:E1130-E1139.
Barona-Gomez, F., Lautru, S., Francou, F.X., Leblond, P., Pernodet, J.L., Challis, G.L. (2006). Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3 (2) and Streptomyces ambofaciens ATCC 23877. Microbiology, 152:3355-3366.
Zawadzka, A.M., Abergel, R.J., Nichiporuk, R., Andersen, U.N., Raymond, K.N. (2009). Siderophore-mediated iron acquisition systems in Bacillus cereus: Identification of receptors for anthrax virulence-associated petrobactin. Biochemistry, 48:3645-3657.
Baghaee Ravari, S., Heidarzadeh, N. (2014). Isolation and characterization of rhizosphere auxin producing Bacilli and evaluation of their potency on wheat growth improvement. Archives of Agronomy and Soil Science, 60:895-905.
Yu, X., Ai, C., Xin, L., Zhou, G. (2011). The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. European Journal of Soil Biology, 47:138-145.
Woo, S.M., Kim, S.D. (2008). Structural Identification of $ Siderophore_ {AH18} $ from Bacillus subtilis AH18, a Biocontrol agent of Phytophthora Blight Disease in Red-pepper. Microbiology and Biotechnology Letters, 36:326-335.Author Info
T.P. Dunyashev1*, G.Yu. Laptev1, E.A. Yildirim2, L.A. Ilina2, V.A. Filippova2, D.G. Tiurina2, A.V. Dubrovin2, N.V. Tarlavin2, S.N. Bikonya2, E.A. Brazhnik2, V.H. Melikidy2 and A.V. Platonov32BIOTROF Ltd., MalinovskayaSt, 8A, 7-N, Pushkin, StPetersburg, 196602, Russian Federation
3Vologda Research Center of the Russian Academy of Sciences, Gorky, St, 56a, Vologda,160014, Russian Federation
Citation: Dunyashev, T.P., Laptev, G.Yu., Yildirim, E.A., Ilina, L.A., Filippova, V.A., Tiurina, D.G., Dubrovin, A.V., Tarlavin, N.V., Bikonya, S.N., Brazhnik, E.A., Melikidy, V.H., Platonov, A.V. (2021). Analysis of the potential associated with the siderophores synthesis in the Bacillus subtilis strain using whole genome sequencing. Ukrainian Journal of Ecology. 11 (7), 116-120.
Received: 04-Aug-2021 Accepted: 18-Sep-2021 Published: 27-Sep-2021
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
