Research - (2023) Volume 13, Issue 2
A novel Niallia nealsonii bacteria degrading phenol isolated from Oum ghellaz lake shore soil in Oran in Algeria
D. Maghnia1,2*, F. Adoudj3 and A. Abdessalem Arezki1Abstract
A new indigenous soil phenol-degrading bacterium strain S2 was successfully isolated from Oum ghellaz lake shore soil in Oran in Algeria. Based on its morphological, physiological and biochemical characteristics, the strain S2 was characterized as a Gram-positive, facultatively anaerobic, forms endospores, and short rod-shaped bacterium that utilizes phenol as a sole carbon and energy source also other phenolic compounds. 16SrDNA sequence analysis revealed that this strain is affiliated to Niallia nealsonii in the group of Firmicutes. The strain was efficient in removing 91.6% of the initial 500 mgL-1 phenol within 48 h, and had a tolerance of phenol concentration as high as 1500 mgL-1. These results indicated that Niallia nealsonii possesses a promising potential in the bioremediation.
Keywords
Biodegradation, Phenol, Niallia nealsonii, Oum ghellaz lake.
Introduction
Phenolic compounds are organic pollutants that are very harmful to both humans and the environment. Indeed, some of them can be highly toxic due to their mutagenic and/or carcinogenic properties (Das et al., 2016). Phenols are produced annually at a rate of 7 million tons in the world. They are discharged by various industries such as oil refineries, coking plants, pharmaceutical industry, chemical industry, etc. (Senthilvelan et al., 2014). Due to its high water solubility, phenol in wastewater reaches downstream water sources and can harm life in aquatic environments (Deng et al., 2018). Due to the limits of physico-chemical treatment which are costly, non-ecological, complex, can lead to the destruction of the texture, the characteristics of the soil, of the waters, the bioremediation by using microorganisms is recognized as a valuable alternative for detoxification and elimination of the toxic substance, due to their cost-effectiveness, respect for the environment and their technological simplicity (Ren et al., 2017). It is an evolutionary method for the removal and degradation of many environmental pollutants, including phenols (Samiml et al.,2020).
Many phenol-degrading microorganisms, including bacteria, yeast and algae have been isolated from environment, among which the bacteria are studied extensively, like Pseudomonas putida (Kumar et al., 2005), Burkholderia sp. (Arora and Jain, 2012), Kocuria sp. (Wu et al., 2018), Acinetobacter sp. (Jiang et al., 2013; Iqbal et al., 2018), Arthrobacter sp. (Wong et al., 2015), Bacillus sp. (Banerjee and Ghoshal, 2010; Hasan and Jabeen, 2015; Iqbal et al., 2018), Halomonas sp. (Haddadi and Shavandi, 2013), Pseudomonas aeruginosa (Hasan et al., 2015), Citrobacter sp. (Deng et al., 2018), Raoultella sp. (Jayachandran et al., 2018) Pseudomonas sp. ATR208 (Sepehr et al., 2019). However, there is no information available regarding phenol degradation activity within the genus Niallia. This paper describes the isolation and identification of a novel phenol-degrading Gram-positive bacterium of Niallia species Oum ghellaz lake shore soil in Oran in Algeria and to determine the kinetics of biodegradation.
Materials and Methods
Chemicals, media and soil sample
All chemicals used were analytical reagents. The minimal salt media (MSM) and Luria-Bertani (LB) media were used in present study. The MSM contained KH2PO4 0.5 g, K2HPO4 0.5 g, CaCl2 0.1 g, NaCl 0.2 g, MgSO4.7H2O 0.5 g, MnSO4.7H2O 0.01 g, FeSO4.7H2O 0.01 g, NH4NO3 1.0 g per liter. The LB media was composed of tryptone 10 g, yeast extract 5 g and NaCl 5 g per liter. Deionized, distilled water was used for the experiments. The soil samples were collected from Oum ghellaz lake shore soil in Oran in Algeria by carefully scraping the soil using a sterile spatula. The soil was sieved under aseptic conditions to a particle size of approximately 2-4 mm, and then 10 g of soil was added to 90 ml of of the minimum medium based on mineral salts (MSM) for enrichment culture.
Isolation and enrichment of aerobic bacteria that degrade phenol
The sample were collected Oum ghellaz lake shore soil, 10 g of soil was inoculated into flasks containing 90 ml of MSM for the enrichment culture. Phenol was supplemented in the media as the sole carbon source, and the various concentrations of phenol were 200, 500, 800, 1100, 1400 and 1700 mgL-1. The enriched culture observed with more biomass (in the flask with 500 mgL-1 phenol) was further transferred into a freshly prepared enrichment media with higher concentrations of phenol (increased from 500 mgL-1 to 1500 mgL-1). The final enriched media were diluted serially and spread on LB agar plates supplemented with phenol (500 mgL-1). The plates were incubated at 30°C and single colonies with morphological differences were selected and streaked on new plates. The resulting isolates were stored at 4°C for further study.
To test the ability of the isolate to use other phenolic compounds as a sole source of carbon and energy, the strain were spread onto MSM plates and the carbon source was supplemented via vapor phase by adding it in the sterile Eppendorf plastic tip (50 μl) and placing the tip in the lid of the Petri dish. After 2-4 days of incubation at 30°C, the plates were screened for the presence of colonies. Growth was confirmed by comparison with control plates without substrate and 20 mM glucose as carbon source, respectively (Djokic, 2011).
Taxonomic identification and characterization of phenol degrading bacteria
One strain capable of achieving high biomass yields with phenol as a sole carbon and energy source were selected for further phenotypically characterization. The morphological properties of the isolated colonies were observed by optical microscopy. The typical physiological and biochemical characteristics of the phenol-degrading bacteria strains, such as Gram’s staining, motility, starch hydrolysis, and gelatinase (kloos et al., 1974) were systematically performed according to Bergey’s manual of determinative of bacteriology (Holt et al., 1998) Indole test, methyl red test were also analyzed (Zeinat et al., 2008).
This strain were also identified by sequence analysis of 16S rRNA genes. The genomic DNA of bacterial strains was extracted by using the Promega Genomic DNA Extraction kit. The quantity and the quality of DNA extracts are monitored by the NanoDrop spectrophotometer (thermo scientific,USA). The extracted DNA was used as template to amplify bacterial 16Sr DNA with universal primers 27F and 1492R (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1392R (5′-ACGGGCGGTGTGTGTRC-3′) with a Biorad cycler thermo-cycler (Biorad, USA). The amplification PCR was performed under the following conditions: An initial denaturation step of 5 min at 94°C was conducted, followed by 35 cycles of 94°C for 30 s, 55°C for 45 s and 72°C for 90s. The procedure was completed with a final elongation step at 72°C for 10 min. The PCR products were sequenced and the sequences were compared with bacterial 16Sr DNA sequences in GenBank (National Centre for Bio-technology Information, Rockville Pike, Bethesda, MD), (http://www.ncbi.nlm.nih.gov/) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Altschul et al., (1997) were constructed using the Molecular Evolutionary Genetics Analysis (MEGA version 6) (Tamura et al., 2011). The reliability of phylogenetic reconstructions was estimated through bootstrap analysis (1000 replicates).
Phenol degradation
The culture of strain was prepared and adjusted to an optical density at 600 nm (OD600) of 1.0, then the final concentration of 2% (v/v) inoculums were inoculated into the flasks containing MSM media with phenol as sole carbon source (katarzyna et al., 2012) the range of phenol concentrations was increased from 100 to 1500 mgL-1. The flasks were incubated at 30°C with 150 rpm for 3 days. Samples were collected periodically to measure the biomass and the phenol degradation. The biomass contents were monitored spectrophotometrically by measuring absorbance at 600 nm. The phenol concentrations were determined by using 4-aminoantipyrine in the colorimetric assay, according to standard methods reported by the (APHA., 2005).
Results and Discussion
Isolation and characterization of phenol-degrading strains
After three weeks of enrichment and one week of strain isolation, a total of 10 isolates were obtained after 24 h growth on the LB agar plates with 100 μL of a 10-5-10-6 fold dilution of enrichment culture. All these stains utilized phenol as the sole carbon source and energy, and 1 of the 10 isolates exhibited more growth in phenol-containing media than the others. The outstanding isolate was named S2 and was applied in the following study. The selected strain were tested for it’s ability to grow on a range of additional 10 phenolic substrates as the sole source of carbon and energy on a solid MSM medium (Table 1). These substrates included o-cresol, m-cresol etc. The isolate S2 were capable of degrading 8 aromatic compounds, including o-cresol, m-cresol, p-cresol, 3,4-DMP, ethylbenzene, benzene, toluene, xylene (Table 1).
| Phenolic compounds | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | Phenol | o-cresol | m- cresol | p-cresol | p-nitro phenol | p-chloro phenol | 3,4-DMP | Benzene | Toluene | Ethyl benzène | Xylene |
| PSA | + | + | + | + | - | - | + | + | +/- | + | +/- |
| PSP | + | + | + | + | - | - | + | + | + | + | + |
| S2 | + | +/- | + | + | - | - | + | + | +/- | + | + |
Table 1. Capability of isolate S2 to use different phenolic compounds.
The strain S2 was a Gram-positive, short rod-shaped bacterium, facultatively anaerobic and forms endospores, colonies on LB agar are irregular, rough, umbonate with undulate edges and beige in colour (Table 2).
| Characteristics | Strain S2 |
|---|---|
| Color of Colony | White |
| Morphology | Short rod |
| Motility | - |
| Gram Straining | + |
| Aerobic Growth | facultatively anaerobic |
| Catalase | + |
| Oxydase | - |
| Glucose | + |
| Mannitiol | - |
| Methyl Red | - |
| Arginine Dihydrolase | - |
| Ornithine Decarboxylase | - |
| Lysine Decarboxylase | - |
| Uree Production | - |
| Indol Production | - |
Table 2. Morphological and biochemical characteristics of S2 strain.
The biochemical characteristics of strain S2 were determined, and biochemical tests showed that Catalase is produced, but oxidase, arginine dihydrolase, lysine and ornithine decarboxylases, uree and indol are not (Table 2). This strain could grow at temperatures range of 30°C-45°C and a wide range of pH 5-11. The optimum growth was at the condition of 30°C-35°C.
The 16S rRNA gene sequences of S2 were sequenced and used to construct a phylogenetic tree for further analysis. The partial sequence of 16S rRNA gene was a continuous stretch of 1180 bp. The similarities between the S2 sequence and the bacterial sequences deposited in the GenBank databases were calculated, and the S2 sequence showed 100% similarity to that of Niallia nealsonii strain MMAPL-X 1 previously named Bacillus nealsonii. The phylogenetic analysis revealed that the strain was classified in the Niallia genera, which belong to the family of Bacillacea from the order Bacillales. Based on neighbor-joining methods, a phylogenetic tree was constructed which indicated that the closest relative of strain S2 was Niallia nealsonii (Fig. 1). Therefore, the strain S2 was identified and affiliated to Niallia nealsonii. 1,393 bp. The obtained sequence was deposited in the Gene-Bank with accession number OQ428241.
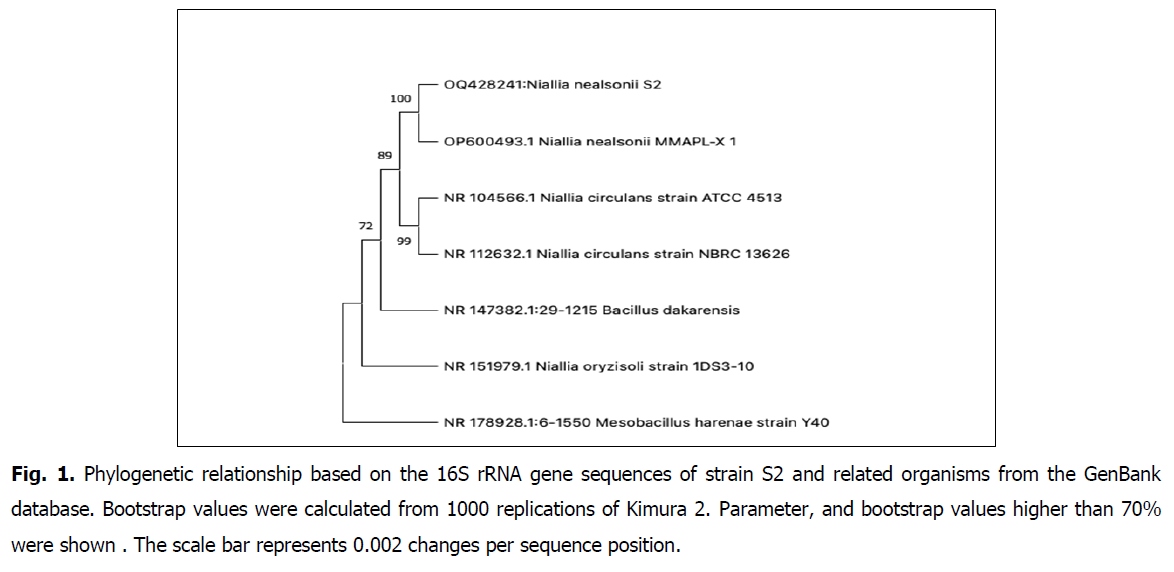
Fig 1. Phylogenetic relationship based on the 16S rRNA gene sequences of strain S2 and related organisms from the GenBank database. Bootstrap values were calculated from 1000 replications of Kimura 2. Parameter, and bootstrap values higher than 70% were shown . The scale bar represents 0.002 changes per sequence position.
Extensive biodegradation studies have described the effectiveness of bacillus species in removing many environmental pollutants from contaminated sites including Bacillus brevis (Arutchelvan et al., 2006), Bacillus cereus (Banerjee and Ghoshal, 2010), Bacillus stearothermophilus (Dong et al., 1992; Gurujey-alakshmi and Oriel, 1989) and Bacillus subtilis (Tam et al., 2006). Banerjee et al., (2010) isolated two Bacillus strains from the oil refinery and exploration sites which could grow on phenol (1000 mgL-1). Nevertheless, no study have been reported on Nialiia nealsonii degrading phenol or aromatic compounds.
The isolation of native microbial species from local polluted environments has been reported to be more adaptive and efficient than non-indigenous microorganisms as biodegraders. Therefore, the isolation of new phenol-degrading bacteria is recommended for the bioremediation of the phenol-contaminated sites in various regions (Liu et al., 2016).
The phenol biodegradation by S2 Niallia nealsonii
The phenol-degradation characteristics and biomass of S2 Niallia nealsonii at various initial concentrations of phenol (100-1500 mgL-1) were determined by monitoring phenol concentration cell growth at OD600 periodically. The maximum biomass and degradation of phenol were observed at the initial phenol concentration of 500 mgL-1 (Fig. 2). An inhibitory effect showed that the biomass growth and the degradation of phenol were declined with the elevated initial phenol concentration higher than 500 mgL-1. The removal rates of phenol were above 79% at the initial phenol concentration ranging from 200 to 600 mgL-1. While there was no growth of bacteria when the initial phenol concentration was higher than 1500 mgL-1. The strain could grow on phenol up to a concentration of 1000 mgL-1 with the degradation rate of 40%.
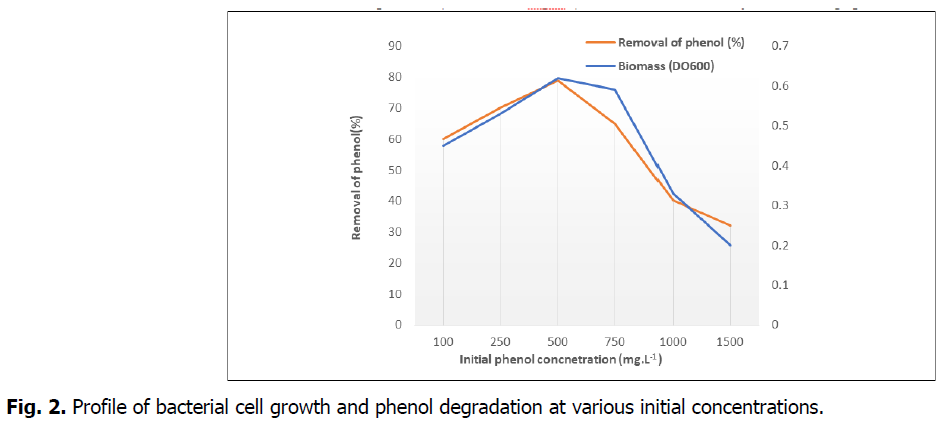
Fig 2. Profile of bacterial cell growth and phenol degradation at various initial concentrations.
The effects of factors such pH values and temperature on the degradation were investigated. bacterial strain could grow within a range of pH 5-11 (Fig. 3a), and the degradation of phenol was above 75% in the range of pH 7-9. The optimum pH for phenol degradation was 7.0. These results showed that the bacterial growth of strain S2 and the degradation of phenol (above 70%) were favored at temperatures of 30°C-35°C (Fig. 3b). The biomass and phenol degradation reached the maximal values at a temperature of 30°C. On the contrary, the phenol degradation declined sharply when the temperature reached 40°C and over. Therefore, the optimal temperature for the growth of strain S2 was 30°C. These growth conditions of Niallia nealsonii. S2 for phenol degradation were similar to those of Bacillus species Strain Bacillus sp. PS11 was isolated and characterized as a strain which utilizes high amount of phenol (up to 1400 mgL-1) in liquid culture without apparent inhibition of growth and it performed well in the initial soil microcosm experiment have been reported by Djokic (2011).
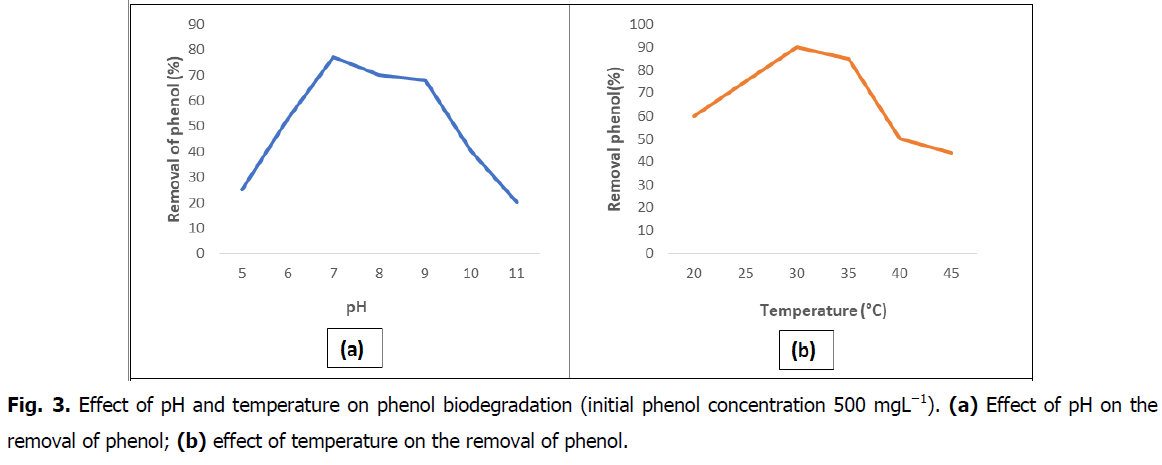
Fig 3. Effect of pH and temperature on phenol biodegradation (initial phenol concentration 500 mgL−1). (a) Effect of pH on the removal of phenol; (b) effect of temperature on the removal of phenol.
Several Bacillus strains have been recognized to grow by using phenol as the sole carbon and Several energy source. However, none study on biodegradation of phenol by Niallia species. In our study the inhibition limit of strain Niallia nealsonii was as high as the phenol concentration of 1500 mgL-1.
Conclusion
In conclusion, a novel bacterial strain capable of degrading phenol was isolated from Oum ghellaz lake shore soil in Oran in Algeria and it was identified as Niallia nealsonii S2 based on the 16S rDNA sequence and the phylogenetic analysis. Niallia nealsonii S2 has the ability to grow in a liquid medium with phenol at different concentrations as the sole carbon and energy and to use other phenolic compounds as substrat (including o-cresol, m-cresol, p-cresol, 3,4-DMP, Ethylbenzène,benzene, toluene, Xylène).
The strain was able to degrade 80% of the initial 500 mgL-1 phenol and grow at the phenol concentration of as high as 1500 mgL-1.
The optimal growth conditions for phenol degradation of strain were at 30°C and pH 7.0. Regarding that native microbial species were more adaptive than non-indigenous microorganisms in polluted environments, their predominance facilitated the bioremediation of the phenol-contaminated environments. Niallia nealsonii S2 isolated may be the used for the bioremediation of the phenol-contaminated environments in Algeria.
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215:403-410.
Google Scholar, Crossref, Indexed at
APHA. (2005). Standard methods for the examination of water and wastewater. American Public Health Association, Washington DC.
Arora, P.K., Jain, R.K. (2012). Metabolism of 2-chloro-4-nitrophenol in a Gram negative bacterium, Burkholderia sp. RKJ 800. PloS One, 7:e38676.
Arutchelvan, V., Kanakasabai, V., Nagarajan, S., Muralikrishnan, V. (2005). Isolation and identification of novel high strength phenol degrading bacterial strains from phenol-formaldehyde resin manufacturing industrial wastewater. Journal of Hazardous Materials, 127:238-243.
Google Scholar, Crossref, Indexed at
Banerjee, A., Ghoshal, A.K. (2010). Phenol degradation by Bacillus cereus: Pathway and kinetic modeling. Bioresource Technology, 101:5501-5507.
Google Scholar, Crossref, Indexed at
Das, N., Chandran, P. (2011). Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnology Research International.
Das, B., Mandal, T.K., Patra, S. (2016). Biodegradation of phenol by a novel diatom BD1IITG-kinetics and biochemical studies. International Journal of Environmental Science and Technology, 13:529-542.
Deng, T., Chen, Y., Liu, B., Laguna, M.P., de la Rosette, J.J., Duan, X., Zeng, G. (2019). Systematic review and cumulative analysis of the managements for proximal impacted ureteral stones. World Journal of Urology, 37:1687-1701.
Djokic, L., Narancic, T., Nikodinovic-Runic, J., Savic, M., Vasiljevic, B. (2011). Isolation and characterization of four novel Gram-positive bacteria associated with the rhizosphere of two endemorelict plants capable of degrading a broad range of aromatic substrates. Applied Microbiology and Biotechnology, 91:1227-1238.
Dong, F.M., Wang, L.L., Wang, C.M., Cheng, J.P., He, Z.Q., Sheng, Z.J., Shen, R.Q. (1992). Molecular cloning and mapping of phenol degradation genes from Bacillus stearothermophilus FDTP-3 and their expression in Escherichia coli. Applied and Environmental Microbiology, 58:2531-2535.
Gurujeyalakshmi, G., Oriel, P. (1989). Isolation of phenol-degrading Bacillus stearothermophilus and partial characterization of the phenol hydroxylase. Applied and Environmental Microbiology, 55:500-502.
Hasan, S.A., Jabeen, S. (2015). Degradation kinetics and pathway of phenol by Pseudomonas and Bacillus species. Biotechnology and Biotechnological Equipment, 29:45-53.
Haddadi, A., Shavandi, M. (2013). Biodegradation of phenol in hypersaline conditions by Halomonas sp. strain PH2-2 isolated from saline soil. International Biodeterioration and Biodegradation, 85:29-34.
Google Scholar, Crossref, Indexed at
Holt, S.G., Kriey, N.R., Sneath, P.H.A., Staley, J.T. (2008). Bergey’s manual of determinative for bacteriology. New York: Williams and Wilkins; 1996. Shoreline. Applied and Environmental Microbiology, 104:251-259.
Iqbal, A., Arshad, M., Hashmi, I., Karthikeyan, R., Gentry, T.J., Schwab, A.P. (2018). Biodegradation of phenol and benzene by endophytic bacterial strains isolated from refinery wastewater-fed Cannabis sativa. Environmental Technology, 39:1705-1714.
Jayachandran, K., Anoop, M., Indu, C.N. (2018). Growth kinetics of alcaligenes sp d2 during phenol biodegradation in mineral salt phenol medium. World Journal of Pharmaceutical Research, 7:1084-92.
Jiang, L., Ruan, Q., Li, R., Li, T. (2013). Biodegradation of phenol by using free and immobilized cells of Acinetobacter sp. BS8Y. Journal of Basic Microbiology, 53:224-230.
Kumar, A., Kumar, S., Kumar, S. (2005). Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochemical Engineering Journal, 22:151-159.
Google Scholar, Crossref, Indexed at
Kloos, W.E., Tornabene, T.G., Schleifer, K.H. (1974). Isolation and characterization of micrococci from human skin, including two new species: Micrococcus lylae and Micrococcus kristinae. International Journal of Systematic and Evolutionary Microbiology, 24:79-101.
Liu, Z., Xie, W., Li, D., Peng, Y., Li, Z., Liu, S. (2016). Biodegradation of phenol by bacteria strain Acinetobacter calcoaceticus PA isolated from phenolic wastewater. International Journal of Environmental Research and Public Health, 13:300.
Google Scholar, Crossref, Indexed at
Ren, L.F., Chen, R., Zhang, X., Shao, J., He, Y. (2017). Phenol biodegradation and microbial community dynamics in extractive membrane bioreactor (EMBR) for phenol-laden saline wastewater. Bioresource Technology, 244:1121-1128.
Google Scholar, Crossref, Indexed at
Senthilvelan, T., Kanagaraj, J., Panda, R.C., Mandal, A.B. (2014). Biodegradation of phenol by mixed microbial culture: an eco-friendly approach for the pollution reduction. Clean Technologies and Environmental Policy, 16:113-126.
Sepehr, S., Shahnavaz, B., Asoodeh, A., Karrabi, M. (2019). Biodegradation of phenol by cold-tolerant bacteria isolated from alpine soils of Binaloud Mountains in Iran. Journal of Environmental Science and Health, Part A, 54:367-379.
Silva, Í.S., dos Santos, E.D.C., de Menezes, C.R., de Faria, A.F., Franciscon, E., Grossman, M., Durrant, L.R. (2009). Bioremediation of a polyaromatic hydrocarbon contaminated soil by native soil microbiota and bioaugmentation with isolated microbial consortia. Bioresource Technology, 100:4669-4675.
Google Scholar, Crossref, Indexed at
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28:2731-2739.
Tam, L.T., Eymann, C., Albrecht, D., Sietmann, R., Schauer, F., Hecker, M., Antelmann, H. (2006). Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environmental Microbiology, 8:1408-1427.
Wu, L., Ali, D.C., Liu, P., Peng, C., Zhai, J., Wang, Y., Ye, B. (2018). Degradation of phenol via ortho-pathway by Kocuria sp. strain TIBETAN4 isolated from the soils around Qinghai Lake in China. PloS One, 13:e0199572.
Wang, Y., Wang, C., Li, A., Gao, J. (2015). Biodegradation of pentachloronitrobenzene by Arthrobacter nicotianae DH19. Letters in Applied Microbiology, 61:403-410.
Zeinat Kamal, M., Nashwa, A.H., Mohamed, A.I., Sherif, E.N. (2008). Biodegradation and detoxification of malathion by of Bacillus thuringiensis MOS-5. Australian Journal of Basic and Applied Sciences, 2:724-732.
Author Info
D. Maghnia1,2*, F. Adoudj3 and A. Abdessalem Arezki12Laboratory of Bio-economy, Food Security and Health, University of Mostaganem Ahmed Ibn Badis, Algeria
3Laboratory of Applied Microbiology, University of Oran Ahmed Ben Bella 1, Algeria
Citation: Maghnia, D., Adoudj, F., Abdessalem Arezki, A. (2023). A novel Niallia nealsonii bacteria degrading phenol isolated from Oum ghellaz lake shore soil in Oran in Algeria. Ukrainian Journal of Ecology. 13:37-43.
Received: 20-Feb-2023, Manuscript No. UJE-23-89692; , Pre QC No. P-89692; Editor assigned: 22-Feb-2023, Pre QC No. P-89692; Reviewed: 07-Mar-2023, QC No. Q-89692; Revised: 13-Mar-2023, Manuscript No. R-89692; Published: 18-Mar-2023, DOI: 10.15421/2023_428
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
